 Funnotate
Funnotate
Loading, please wait.
Annotate protein or nucleotide sequences using LIS legume resources, and identify homologous gene families.
(Or search for gene families using this functional keyword search.)
Because this service involves several computationally intensive searches (see pipeline description below), results can take from several minutes to several hours, depending on the size of your query. Thanks for your patience.
Upload your protein or nucleotide FASTA sequence(s) (max. 100 kbp)
Annotation pipeline:
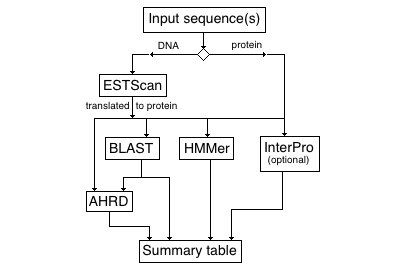
1. ESTScan to translate nucleotide sequences (if needed)
2. BLAST alignment to reference gene databases (soybean, Medicago, Arabidopsis)
3. (optional) InterProScan (methods: TIGRFAM, SMART, SUPERFAMILY, Gene3D, PIRSF, Pfam, Coils)
4. HMMer for assigning gene family
5. AHRD (Automated Human Readable Descriptions) for best-hit extraction
Summary table results include: Legume Federation gene family, AHRD descriptor, best BLAST hits, GO and InterPro ID (the latter two only if InterProScan is run)
Complete output files for each analysis are also provided.
Note that the gene families which are searched are the families displayed on LIS, i.e. those having at least one member which is an LIS species.
Enter keyword(s) to search for functional descriptions in gene families.
Use quotes to search for phrases ("cysteine methyl"),
* for partial matches (cyst*),
OR (AND) to search for either (both) of two terms (cysteine OR methyl; cysteine AND methyl),
NOT to exclude a term (cysteine AND NOT methyl).
(Or go to the Funnotate home page to annotate your sequence(s) and identify homologous gene families.)
Gene Family Selection
Click a gene family to display its phylogram.
Please verify that this is correct before starting your job - or start over
1GF Score = full-sequence E-value from hmmscan
2BLAST Score: E-value is displayed, however best hit is chosen using bit score
3AHRD Quality Scores are as follows:
The AHRD quality code consists of a three-character string, where each character is either * if the respective criteria are met or _ otherwise. The meaning by position is as follows:
1 - Bit score of the BLAST result is over 50 and e-value is better than e-10
2 - Overlap of the BLAST result is at least 60%
3 - Top token score of assigned descriptor is at least 0.5
Note that the Best BLAST Hit may differ from the AHRD BLAST Hit because AHRD factors in the information content of the sequence description.
For further explanation of these codes and the AHRD algorithm, see AHRD Documentation.